High-Speed 3D Tracking of Chlamydomonas with Phase-Contrast Microscope
Summary
We have applied our microorganism tracking system to assess flagella mechanism of chlamydomonas, collaborating with T. Yamano and Prof. M. Kikkawa in Graduate School of Medicine and Faculty of Medicine, University of Tokyo.
Recently, flagella and cilia have gathered attension since their important role in mammal cells and relation to genetic diseases of human were revealed. Chlamydomonas is one of model organism of flagella research. The purpose of our research is to track a freely swimming indivisual chlamydomonas cell to observe its flagella and whole body movement and to study flagella mechanism.
Flagella of Chlamydomonas are very small structure with length of 10-15 micro meter, thickness of 200-300 nm. Special microscopic technique such as phase-contrast microscopy is essential to observe such small target. However, formally developed high-speed autofocusing algorithm named Depth From Diffraction (DFDi) could not be applied to the phase-contrast microscope.
A new method was developed to achieve high-speed autofocus with the phase-contrast microscope, based on the DFDi technique. This new method estimates depth position of individual chlamydomonas from single image by assuming the linear relationship between the radial intensity profile of the cell image and its depth position.
Though the estimated depth includes considerable error due to the linear assumption, the estimated depth had sufficient precision for the visual tracking, because of focusing control can be achieved by coarse depth resolution. In the minimum case, only three states, far from focus, in focus, and near from focus, are sufficient to achieve focus tracking, since the target is just needed to be shifted continuously in the direction of in focus if the state is not in focus.
Experimental results showed that we successfully achieved continuous stable 3D tracking of swimming Chlamydomonas with quality sufficient for assessing flagella movement, indicating the feasibility of our system.
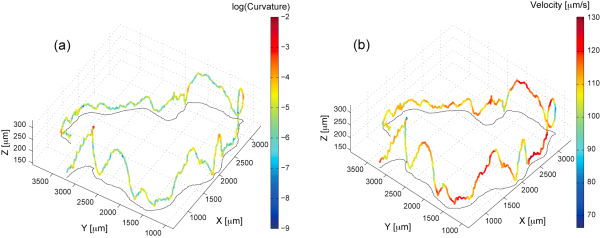
Figure 1. Three-dimensional trajectory of a free swimming chlamydonomas, (a) showing the curvature, and (b) showing the velocity.



