High-Speed DFDi Algorithm for Multiple Cells
Summary
We proposed a new autofocusing method for microbiological specimens, such as cells, using depth information included in their diffraction pattern. This method was named as Depth From Diffraction (DFDi).
The previously developed image processing algorithm of DFDi could estimate depth of a cell only when single cell is included in a field of view. However, it's common case when multiple cells exist in the field of view, considering real applications such as scanning microscopy and automated inspections. Thus, we developed a new image processing algorithm for DFDi that can estimate depth of multiple cells.
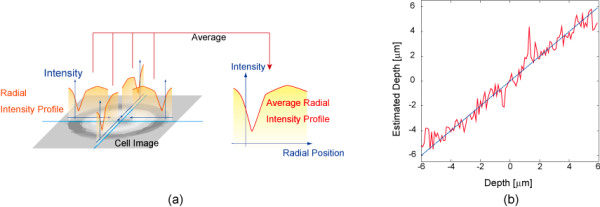
This algorithm recognizes boundaries of each cell in the acquired image first. Then it extracts radial intensity profiles which is image intensity profile along perpendicular direction to the cell boundary. A schematic figure of this process is shown in the following figure (a). Finally, it estimates the depth of cells from the extracted radial intensity profiles.
This algorithm was applied to yeast cell. Figure (b) shows it successfully estimated its depth with an error of 1 um. It also applied to high-speed autofocusing of multiple yeast cells. This result is shown in High-Speed Scanning Microscope by Depth From Diffraction (DFDi) Method.
Reference
- Soshiro Makise, Hiromasa Oku, and Masatoshi Ishikawa: Serial Algorithm for High-speed Autofocusing of Cells using Depth From Diffraction (DFDi) Method, 2008 IEEE International Conference on Robotics and Automation (ICRA 2008) (Pasadena, 2008.5.23)/Conference Proceedings, pp.3124-3129 [PDF (4.1M)] *IEEE
*IEEE © 2008 IEEE. Personal use of this material is permitted. However, permission to reprint/republish this material for advertising or promotional purposes or for creating new collective works for resale or redistribution to servers or lists, or to reuse any copyrighted component of this work in other works must be obtained from the IEEE.



