Visualization and Decoding of External Stimuli Perceived by Living Microorganisms
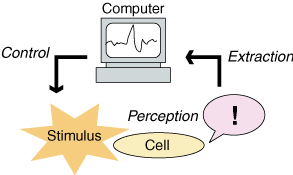
In this paper, we propose a novel microsensing scheme in which intrinsic sensing capability in microorganisms is utilized. Extraction of signals inside the cell and decoding of stimuli received would be an interesting challenge toward development of novel biomimetic sensors, or for potential utilization of a living microorganism itself as a microrobot. As a prototype, we focus on the mechanosensory process in Paramecium cells. When a mechanical stimuli is applied, calcium ion concentration in the cell rises. By visualizing the calcium level rise by using calcium fluorescent indicators and input intensity data into PCs, we can extract the sensation perceived by the cell. A simple experiment was performed in vivo and contact sensation was successfully extracted and decoded. It can be applied for on-board sensors in cells as microrobots in future works. Technology to link living cells and computers would lead to the Cell Machine Interface, to come next after the forthcoming brain machine interface (BMI) paradigm.
Summary
The sensing ability of living things are superior to most of the existing artificial microsensors. It would be of great benefit and interest to utilize such sophisticated capabilities built in living cells in vivo. As illustrated in figures below, extraction, decoding, and feedback of the sensation received by microorganisms by computers would be an interesting challenge toward development of novel biomimetic sensors, or for potential utilization of a living microorganism itself as a sensor, coupled with robotic maneuvers of living cells that the authors have achieved.
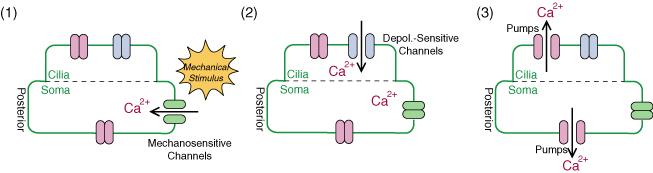
As an example, we introduce Paramecium caudatum, a kind of unicellular protozoa, which we have utilized for microrobotic application. A Paramecium cell detects a contact stimuli and swims back so as to avoid an obstacle. During this behavior, the calcium ion concentration rises inside the cell. In other words, the stimulus information is coded as a calcium signal. If we can extract this mechanosensation signal and decode it, we can estimate what the cell perceived. For extraction of the signal, some visualization methods would be effective. For example, calcium indicators can visualize the intracellular calcium concentration, as illustrated below.
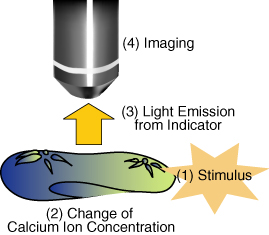
As a prototype, we considered visualization of contact stimuli using Paramecium caudatum cells. When a paramecium cell receives a contact stimulus, a rise in calcium concentration occurs inside the cell, which can be visualized by calcium indicators. We introduced an indicator into cells by microinjection, visualize the signal transduction, and estimate the timings of when the cell detected contact stimuli.
Experimental Results
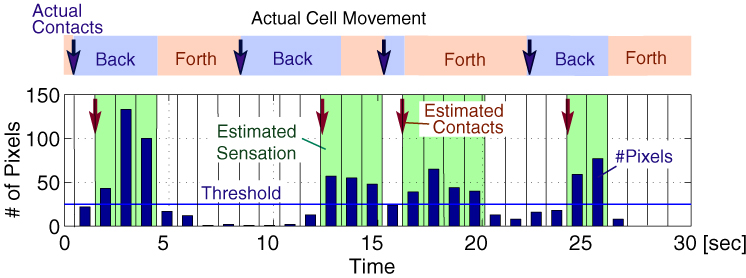
The figure below shows an example of the captured image sequence of the detected fluorescence. After stimulus application, one can find that the calcium level increased rapidly and then decayed slowly.
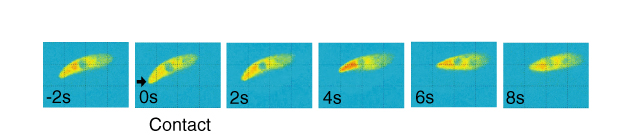
From the intensity of the pseudocolored image sequences, we retrieved the information on whether or not the head of the cell made a contact, i.e., on/off 1 bit information. Using calcium level data, we estimated the sensation the cell perceived, and the timings of contact. The estimation was performed off-line by MATLAB. Four contacts occurred in 30s were successfully estimated with a few seconds of delays.
Reference
- Anchelee Davies, Naoko Ogawa, Hiromasa Oku, Koichi Hashimoto and Masatoshi Ishikawa. Visualization and Estimation of Contact Stimuli using Living Microorganisms. 2006 IEEE International Conference on Robotics and Biomimetics (ROBIO 2006) (Kunming, China, 18 Dec 2006) / Proceedings, pp. 445-450, Dec. 2006. (Best Paper in Biomimetics) [PDF (317K)] *IEEE
Corresponding author: Naoko Ogawa
(Naoko_Ogawa )
)



